
The new version of the CAESAR models, that will be made freely available soon, will include some new features, helpful both to improve usability of the software and to obtain more reliable predictions.
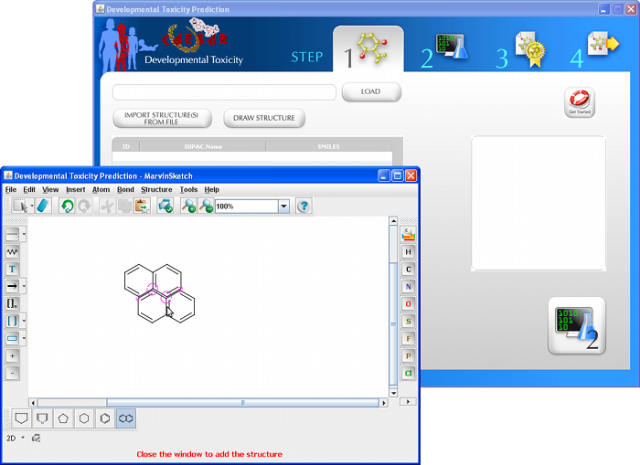
Importing and loading structures
With CAESAR 2.0 you will be able to submit molecules in many different ways:
- Import multiple molecules in sdf and/or smi file format
- Draw structure using the internal editor*
- Enter molecules one-by-one using SMILES format, molecule name (IUPAC) and CAS number

* The molecule editor included is part of the ChemAxon Marvin package.
More info at http://www.chemaxon.com/marvin/help/index.html
The image on the right, as well as all the images used as preview in this page, refers to the stand-alone version of CAESAR software, which anticipate most of the new features of CAESAR 2.0.
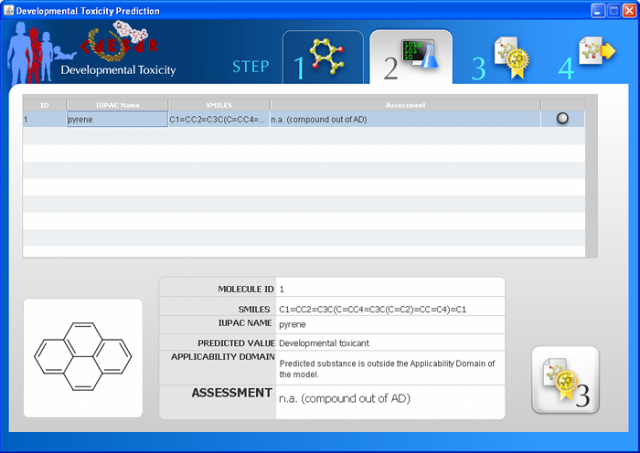
Results reliability & model applicability domain
According to REACH, user should check if "the substance is included in the applicability domain of the model", for a correct use of the (quantitative) structure-activity relationship (Q)SAR model.
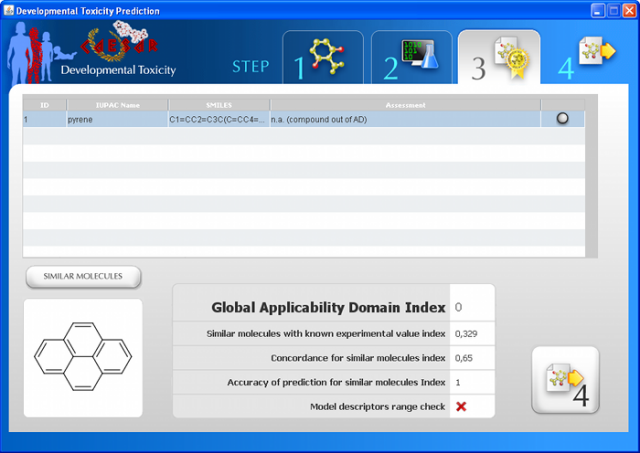
CAESAR 2.0 integrates a tool to assess the applicability domain (AD), through quantitative and visual ways. This free tool is based on:
- Chemiometric check,
- Fragments for outliers,
- Similarity index,
- Prediction Concordance with experimental values,
- Prediction Accuracy of the prediction,
- Uncertainty of the prediction.
Thus, the CAESAR's tool is based not only on the chemical information, as the typical AD tools, but also on toxicity results.
Furthermore, it uses not only a priori data (from the chemical structure), but also a posteriori ones, from the results of the models. This tool proved to discriminate cases where QSAR can be applied, and cases were there may be problems.


 WARNING: the MUTAGENICITY model could classify a compound as mutagen even if it is formally out of the applicability domain. This behaviour is normal for this model and it is related to the use of structural alerts [ref.].
WARNING: the MUTAGENICITY model could classify a compound as mutagen even if it is formally out of the applicability domain. This behaviour is normal for this model and it is related to the use of structural alerts [ref.].


Apart from this new feature, the user could still compare the prediction obtained for a molecule with those obtained for its six most similar molecules founded in the dataset used to build the model, as in the current version of CAESAR models.

Most of the new features that will be present in the new CAESAR v2.0 software have been integrated in the stand-alone models for Developmental Toxicity and Mutagenicity
The stand-alone version of CAESAR can be downloaded for free and have been developed to work off-line. The CAS number input method is not available for this version of the software.
If you need help using the software, you can use the video tutorials we have prepared, that will guide you step-by-step explaining exactly how to use the CAESAR stand-alone software.
DOWNLOAD NOW CAESAR v2.0
Download the stand-alone version of the CAESAR v2.0 QSAR model. This version could be run locally with no internet connection needs.
Stand-alone version of the other CAESAR models will be available in the next future.
DEVELOPMENTAL TOXICITY
SIZE: ~100 Mbyte
MUTAGENICITY
SIZE: ~156 Mbyte
COMMON PROBLEMS AND SOLUTIONS
Problem importing structures using "import structure(s) from file".
Software requirements / specs:
-
- Needs JAVA™ 1.5 or higher to run. Click here to download.
- The stand-alone version works on 32bit systems only.

If you need any information and/or support, please contact: coord@caesar-project.eu.
The CAESAR application has been developed by:
- Davide Bigoni (Bigoni Consulting) - Client/Server infrastructure and services
- Alberto Manganaro (IRFMN) - Implementation of Models, Molecular Descriptors and Applicability Domain
- Antonio Cassano (IRFMN) - Implementation of Stand-alone versions
- Todd Martin (US EPA) - Implementation of Molecular Descriptors
- Alexandros Spiliopoulos (University of Patras) - Contribution to the implementation of Developmnetal Toxicity stand-alone
- Aris Avlonitis (University of Patras) - Contribution to the implementation of Developmnetal Toxicity stand-alone
- Adriana Gomez Delgado (AGDesigner) - Graphic layout
- Vittorio Castiglioni (IRFMN) - Graphic contribution for the stand-alone
All the models implemented have been developed by the CAESAR consortium.







No comments:
Post a Comment